Immuno-histochemical image analysis
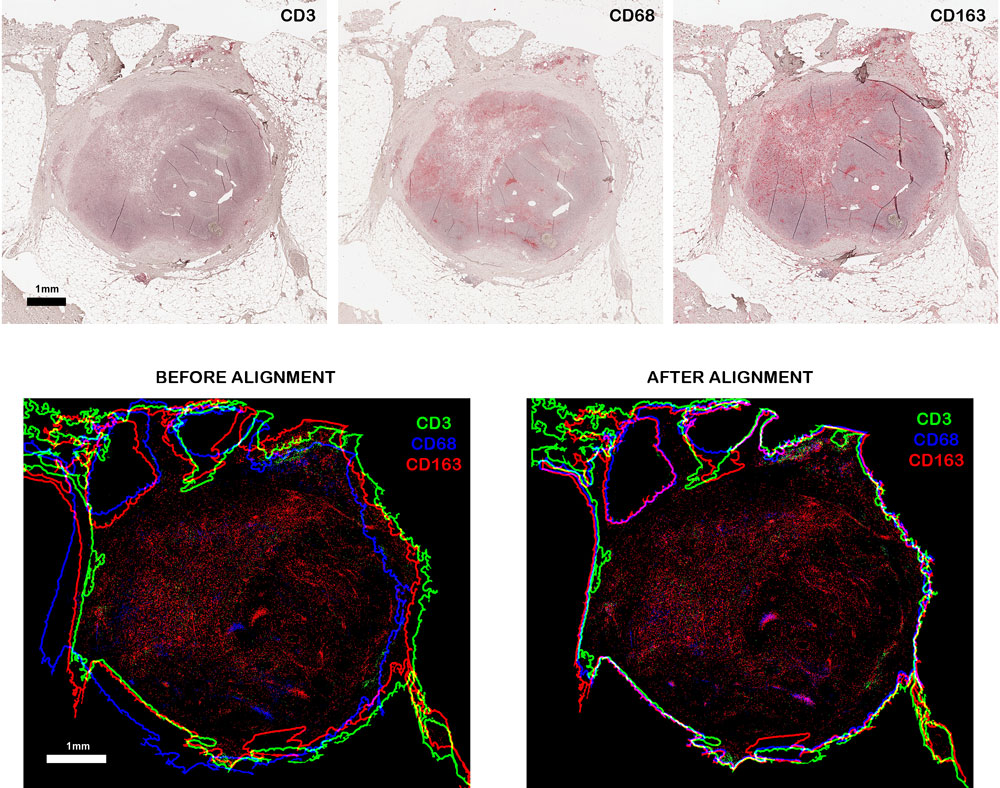
Biomarkers are automatically segmented by a stacked classifier designed to achieve promising results when the training datasets have low cardinality. The extracted markers are quantified by computing their density in the automatically segmented tissue area. Co-occurrence analysis is performed by firstly registering the serialized slices and then expressing co-occurrence through novel measures. The innovative image registration technique, developed to align tissue sections, maximize a function measuring the superimposition of both tissue edges and colors of not marked tissue areas.
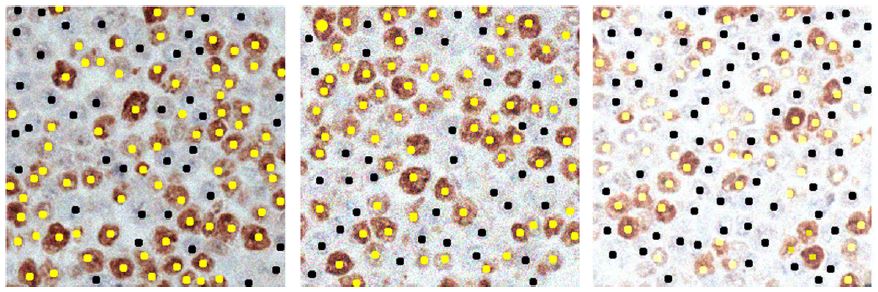
When the marker is the ki67 marker, the ki67 index is automatically estimated by automatically identifying and counting the percentage of tumor cells positive to ki67 with respect to all the tumor cells. The identification of isolated cells and separation of clustered cells is performed by two consecutive multiscale algorithms.
The work has been mainly developed with Consorzio M.I.A. at the University of Milano-Bicocca (Monza, Milan) and Istituto Tumori of Milan (Milan). The work has been part of the research works of the following projects:
- "Early DEtection and Risk Assessment" (EDERA) project, funded by AIRC - Foundation for cancer research.
- "Scaling-up biodegradable nanomedicines for multimodal for multimodal precision cancer immunotherapy" (PRECIOUS) H2020-EU.2.1.2. project.
- A randomized trial in cooperation with Italian Sarcoma Group, Spanish Sarcoma Group, French Sarcoma Group. The trial is entitled: Localized High-Risk Soft Tissue Sarcomas of the Extremities and Trunk Wall in Adults: an Integrating Approach Comprising Standard vs Histotype Tailored NeoAdjuvant Chemotherapy (ISG-STS 10-01).
Publications
B. Barricelli, E. Casiraghi, J. Gliozzo, V. Huber, B. Leone, A. Rizzi and B. Vergani. ki67 nuclei detection and ki67-index estimation: a novel automatic approach based on human vision modeling. BMC Bioinformatics, Springer 20(1), 2019.
V. Huber, V. Vallacchi, V. Fleming, X. Hu, A. Cova, M. Dugo, E. Shahaj, R. Sulsenti, E. Vergani, P. Filipazzi and Others. Tumor-derived microRNAs induce myeloid suppressor cells and predict immunotherapy resistance in melanoma. The Journal of clinical investigation, Am Soc Clin Investig 128(12), 2018.
E. Casiraghi, V. Huber, M. Frasca, M. Cossa, M. Tozzi, L. Rivoltini, B. Leone, A. Villa and B. Vergani. A novel computational method for automatic segmentation, quantification and comparative analysis of immunohistochemically labeled tissue sections. BMC Bioinformatics, BioMed Central 19(10), 2018.
E. Casiraghi, M. Cossa, V. Huber, M. Tozzi, L. Rivoltini, A. Villa and B. Vergani. MIAQuant, a novel system for automatic segmentation, measurement, and localization comparison of different biomarkers from serialized histological slices. European journal of histochemistry: EJH, PAGEPress 61(4), 2017.
 AnacletoLAB
AnacletoLAB